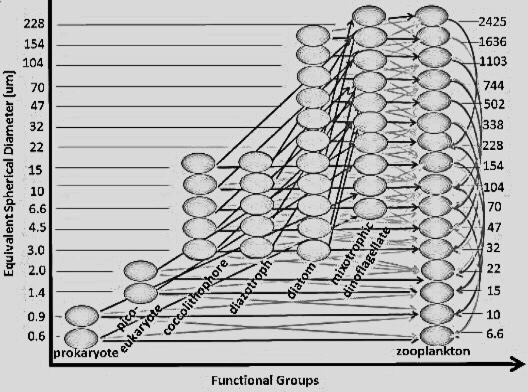
The Simons Collaboration on Computational Biogeochemical Modeling of Marine Ecosystems (CBIOMES) seeks to develop and apply quantitative models of the structure and function of marine microbial communities at seasonal and basin scales.
Latest News

February 2026 CBIOMES e-meeting Scott Pollara (Dalhousie)
“Low proteomic investment in carbon fixation facilitates the success of Phaeocystis in the Labrador Sea” Please note access to this page is restricted to CBIOMES associates. (more…)

February 2026 CBIOMES e-meeting Ioannis Tsakalakis (Hellenic Center for Marine Research, Greece)
”Phytoplankton diel carbon storage enhances trophic transfer and ocean carbon export.” Please note access to this page is restricted to CBIOMES associates. (more…)

An Energy-Based Perspective on Microbial Food Web Structure
New model from CBIOMES researchers reveals how microbes self‑organize to maximize energy flow. (more…)
Latest Publications
Mattern, Jann Paul, Stephanie Dutkiewicz, Jordyn E. Moscoso, Christopher A. Edwards (2026), Zooplankton grazing and nutrient supply control the emergence of large diatoms in coastal upwelling systems: Insights from a regional ecosystem model, Limnology and Oceanography, doi: 10.1002/lno.70332
Jabre, Loay J., Elden Rowland, J. Scott P. McCain, Erin M. Bertrand (2026), Deep Proteomic Profiles of the Antarctic Diatom Fragilariopsis Cylindrus Under Varying Iron and Manganese Conditions, Proteomics, doi: 10.1002/pmic.70109
Williams, R.G., Brown, P.J., Takano, Y. et al. (2026), The biogeochemical transport by the Gulf Stream. Commun Earth Environ, doi: 10.1038/s43247-025-03118-y [Gaël Forget]