CBIOMES members please log in to access. Password issues contact cbiomesweb@gmail.com
April 2024 CBIOMES e-meeting Mick Follows (MIT)
CBIOMES members please log in to access. Password issues contact cbiomesweb@gmail.com
A Marine Symbiosis Allows a Better Understanding of Our Cells Evolution
Human cells, as well as cells of animals, plants, fungi, and other eukaryotic organisms, originally emerged hundreds of millions of years ago through the symbiotic association of some primitive bacteria that, until then, had lived independently. This represented an unprecedented leap in the complexity of life, where some bacteria, after having resided within cells for a long time, eventually transitioned into becoming organelles of these cells. This transition allowed for the compartmentalization and control of bacterial-derived functions within the eukaryotic cell. CBIOMES Mick Follows contributes to a new paper in the journal Cell.
Continue reading “A Marine Symbiosis Allows a Better Understanding of Our Cells Evolution”
NEW CBIOMES PUBLICATION
Ying-Yu Hu, Andrew J. Irwin , Zoe V. Finkel (2024), An improved method to quantify bulk carbohydrate in marine planktonic samples, Limnology and Oceanography Methods, doi: 10.1002/lom3.10614
Get the PDF [Requires login]
NEW CBIOMES PUBLICATION
Stephanie Dutkiewicz, Christopher L. Follett, Michael J. Follows, Fernanda Henderikx-Freitas, Francois Ribalet, Mary R. Gradoville, Sacha N. Coesel, Hanna Farnelid, Zoe V. Finkel, Andrew J. Irwin, Oliver Jahn, David M. Karl, Jann Paul Mattern, Angelicque E. White, Jonathan P. Zehr, Virginia Armbrust (2024), Multiple biotic interactions establish phytoplankton community structure across environmental gradients, Limnology and Oceanography, doi: 10.1002/lno.12555
Get the PDF [Requires login]
NEW CBIOMES PUBLICATION
Francisco M. Cornejo-Castillo, Keisuke Inomura, Jonathan P. Zehr, Michael J. Follows (2024), Metabolic trade-offs constrain the cell size ratio in a nitrogen-fixing symbiosis, Cell, doi: 10.1016/j.cell.2024.02.016
Get the PDF [Requires login]
March 2024 CBIOMES e-meeting Leonhard Lücken (UOL)
CBIOMES members please log in to access. Password issues contact cbiomesweb@gmail.com
March 2024 CBIOMES e-meeting Chris Hill (MIT)
CBIOMES members please log in to access. Password issues contact cbiomesweb@gmail.com
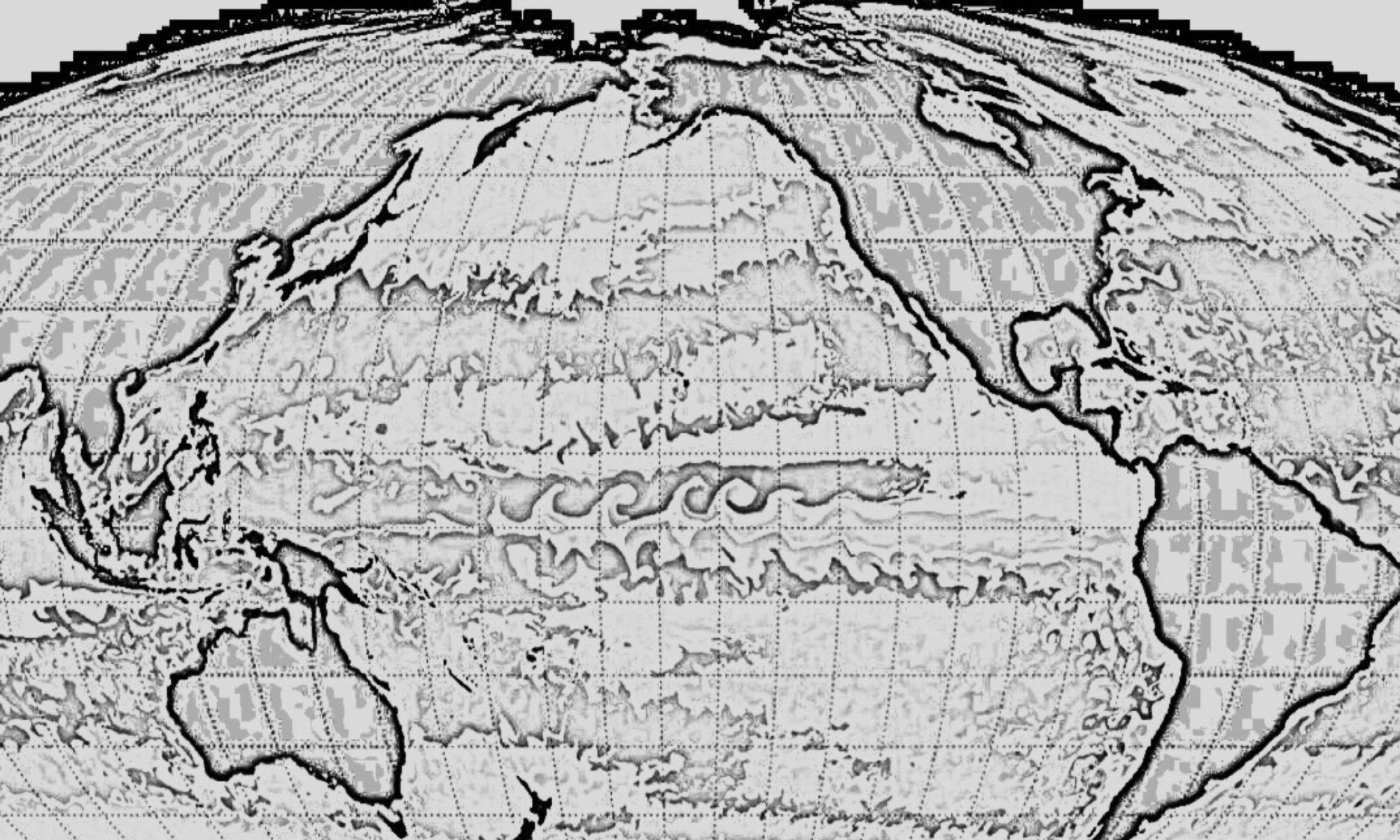
NEW CBIOMES PUBLICATION
Alexandra Jones-Kellett and Michael J. Follows (2024), A Lagrangian coherent eddy atlas for biogeochemical applications in the North Pacific Subtropical Gyre, Earth System Science Data, doi: 10.5194/essd-16-1475-2024
Get the PDF [Requires login]
February 2024 CBIOMES e-meeting Scott McCain (MIT)
CBIOMES members please log in to access. Password issues contact cbiomesweb@gmail.com